It looks like the dust may be settling on the long-running saga of the Fredrickson et al. studies of genomics and well-being, and the Brown et al. reanalyses of the same. We have probably arrived at the end of the discussion in the formal literature. Both sides (of course) think they have won, but the situation on the ground probably looks like a bit of a mess to the casual observer.
I last blogged about this over two years ago. Since then, Fredrickson et al. have produced a second article, published in PLoS ONE, partly re-using the data from the first, and claiming to have found "the same results" --- except that their results were also different (read the articles and decide for yourself) --- with a new mathematical model. We wrote a reply article, which was also published in PLoS ONE. Dr. Fredrickson wrote a formal comment on our article, and we wrote a less-formal comment on that.
I could sum up all of the above articles and comments here, but that would serve little purpose. All of the relevant evidence is available at those links, and you can evaluate it for yourself. However, I thought I would take a moment here to write up a so-far unreported aspect of the story, namely how Fredrickson et al. changed the archived version of one of their datasets without telling anybody.
In the original version of the GSE45330 dataset used in Fredrickson et al.'s 2013 PNAS article, a binary categorical variable, which should have contained only 0s and 1s, contained a 4. This, of course, turned it into basically a continuous variable when it was thrown as a "control" into the regressions that were used to analyse the data. We demonstrated that fixing this variable caused the main result of the 2013 PNAS article --- which was "supported" by the fact that the two bars in Figure 2A were of equal height but opposite sign --- to break; one of the bars more than halved in size.(*) For reasons of space, and because it was just a minor point compared to the other deficiencies of Fredrickson et al.'s article, this coding error was not covered in the main text of our 2014 PNAS reply, but it was handled in some detail in the supporting information.
Fredrickson et al. did not acknowledge their coding error at that time. But by the time they re-used these data with a new model in their subsequent PLoS ONE article (as the "Discovery" sample, which was pooled with the "Confirmation" sample to make a third dataset), they had corrected the coding error, and uploaded the corrected version to the GEO repository, causing the previous version to be overwritten without a trace. This means that if, today, you were to read Fredrickson et al.'s 2013 PNAS article and download the corresponding dataset, you would no longer be able to reproduce their published Figure 2A; you would only be able to generate the "corrected"(*) version.
The new version of the GSE45330 dataset was uploaded on July 15, 2014 --- a month after our PNAS article was accepted, and a month before it was published. When our article appeared, it was accompanied by a letter from Drs. Fredrickson and Cole (who would certainly have received --- probably on the day that our article was accepted --- a copy of our article and the supporting information, in order to write their reply), claiming that our analysis was full of errors. Their own coding error, which they must have been aware of because /a/ we had pointed it out, and /b/ they had corrected it a month earlier, was not mentioned.
Further complicating matters is the way in which, early in their 2015 PLoS ONE article, Fredrickson et al. attempted to show continuity between their old and new samples in their Figure 1C. Specifically, this figure reproduced the incorrect bars from their PNAS article's Figure 2A (i.e., the bars produced without the coding error having been corrected). So Fredrickson et al. managed to use both the uncorrected and corrected versions of the data in support of their hypotheses, in the same PLoS ONE article. I would like to imagine that this is unprecedented, although very little surprises me any more.
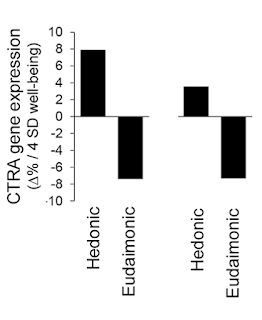
We did manage to get PLoS ONE to issue a correction for the figure problem. However, this only shows the final version of the image, not "before" and "after", so here, as a public service, is the original (left) and the corrected version (right). As seems to be customary, however, the text of Fredrickson et al.'s correction does not accept that this change has any consequences for the substantive conclusions of their research.(*)
Alert readers may have noticed that this correction leaves a problem with Fredrickson et al.'s 2013 PNAS article, which still contains the uncorrected Figure 2A, illustrating the authors' (then) hypotheses that hedonic and eudaimonic well-being had equal and opposite parts to play in determining gene expression in the immune system:
There seems to be one remaining question, which is exactly how unethical it was for the alterations to the dataset to have been made. We made a complaint to the Office of Research Integrity, and it went nowhere. It could be argued, I suppose, that the new version of the data was better than the old one. But we certainly didn't feel that Drs. Fredrickson and Cole had acted in an open and transparent manner. They read our article and supporting information, saw the coding error that we had found, corrected it without acknowledging us, and then published a letter saying that our analyses were full of errors. I find this, if I may use a little British understatement for a moment, to be "not entirely collegial". If this is the norm when critiques of published work are submitted through the peer-review system, as psychologists were recently exhorted to do by a senior figure in the field, perhaps we should not be surprised when some people who discover problems in published articles decide to use less formal methods to comment.
(*) Running through this entire post, of course, is the assumption that the reader has set aside for the moment our demonstration of all of the other flaws in the Fredrickson et al. articles, including the massive overfitting and the lack of theoretical coherency. Arguably, those flaws make the entire question of the coding error moot, since even the "corrected" version of the figures very likely fails to correspond to any real effect. But I think it's important to look at this aspect of the story separately from all of the other noise, as an example of how difficult it can be to get even the most obvious errors in the literature corrected.
I last blogged about this over two years ago. Since then, Fredrickson et al. have produced a second article, published in PLoS ONE, partly re-using the data from the first, and claiming to have found "the same results" --- except that their results were also different (read the articles and decide for yourself) --- with a new mathematical model. We wrote a reply article, which was also published in PLoS ONE. Dr. Fredrickson wrote a formal comment on our article, and we wrote a less-formal comment on that.
I could sum up all of the above articles and comments here, but that would serve little purpose. All of the relevant evidence is available at those links, and you can evaluate it for yourself. However, I thought I would take a moment here to write up a so-far unreported aspect of the story, namely how Fredrickson et al. changed the archived version of one of their datasets without telling anybody.
In the original version of the GSE45330 dataset used in Fredrickson et al.'s 2013 PNAS article, a binary categorical variable, which should have contained only 0s and 1s, contained a 4. This, of course, turned it into basically a continuous variable when it was thrown as a "control" into the regressions that were used to analyse the data. We demonstrated that fixing this variable caused the main result of the 2013 PNAS article --- which was "supported" by the fact that the two bars in Figure 2A were of equal height but opposite sign --- to break; one of the bars more than halved in size.(*) For reasons of space, and because it was just a minor point compared to the other deficiencies of Fredrickson et al.'s article, this coding error was not covered in the main text of our 2014 PNAS reply, but it was handled in some detail in the supporting information.
Fredrickson et al. did not acknowledge their coding error at that time. But by the time they re-used these data with a new model in their subsequent PLoS ONE article (as the "Discovery" sample, which was pooled with the "Confirmation" sample to make a third dataset), they had corrected the coding error, and uploaded the corrected version to the GEO repository, causing the previous version to be overwritten without a trace. This means that if, today, you were to read Fredrickson et al.'s 2013 PNAS article and download the corresponding dataset, you would no longer be able to reproduce their published Figure 2A; you would only be able to generate the "corrected"(*) version.
The new version of the GSE45330 dataset was uploaded on July 15, 2014 --- a month after our PNAS article was accepted, and a month before it was published. When our article appeared, it was accompanied by a letter from Drs. Fredrickson and Cole (who would certainly have received --- probably on the day that our article was accepted --- a copy of our article and the supporting information, in order to write their reply), claiming that our analysis was full of errors. Their own coding error, which they must have been aware of because /a/ we had pointed it out, and /b/ they had corrected it a month earlier, was not mentioned.
Further complicating matters is the way in which, early in their 2015 PLoS ONE article, Fredrickson et al. attempted to show continuity between their old and new samples in their Figure 1C. Specifically, this figure reproduced the incorrect bars from their PNAS article's Figure 2A (i.e., the bars produced without the coding error having been corrected). So Fredrickson et al. managed to use both the uncorrected and corrected versions of the data in support of their hypotheses, in the same PLoS ONE article. I would like to imagine that this is unprecedented, although very little surprises me any more.
We did manage to get PLoS ONE to issue a correction for the figure problem. However, this only shows the final version of the image, not "before" and "after", so here, as a public service, is the original (left) and the corrected version (right). As seems to be customary, however, the text of Fredrickson et al.'s correction does not accept that this change has any consequences for the substantive conclusions of their research.(*)
Alert readers may have noticed that this correction leaves a problem with Fredrickson et al.'s 2013 PNAS article, which still contains the uncorrected Figure 2A, illustrating the authors' (then) hypotheses that hedonic and eudaimonic well-being had equal and opposite parts to play in determining gene expression in the immune system:
But as we have seen, the corrected version of the figure shows a considerable difference between the two bars, representing hedonic and eudaimonic well-being, especially if one considers that the bars represent log-transformed numbers. This implies that the 2013 PNAS article is now severely flawed(*); Figure 2A needs to be replaced, as does the claim about the opposite effects of hedonic and eudaimonic well-being. We contacted PNAS, asking for a correction to be issued, and were told that they consider the matter closed. So now, both the corrected and uncorrected figures are in the published literature, and two different and contradictory conclusions about the relative effects of hedonic and eudaimonic well-being on gene expression are available to be cited, depending on which fits the narrative at hand. Isn't science wonderful?CTRA gene expression varied significantly as a function of eudaimonic and hedonic well-being (Fig. 2A). As expected based on the inverse association of eudaimonic well-being with depressive symptoms, eudaimonic well-being was associated with down-regulated CTRA gene expression (contrast, P = 0.0045). In contrast, CTRA gene expression was significantly up-regulated in association with increasing levels of hedonic well-being (p. 13585)
There seems to be one remaining question, which is exactly how unethical it was for the alterations to the dataset to have been made. We made a complaint to the Office of Research Integrity, and it went nowhere. It could be argued, I suppose, that the new version of the data was better than the old one. But we certainly didn't feel that Drs. Fredrickson and Cole had acted in an open and transparent manner. They read our article and supporting information, saw the coding error that we had found, corrected it without acknowledging us, and then published a letter saying that our analyses were full of errors. I find this, if I may use a little British understatement for a moment, to be "not entirely collegial". If this is the norm when critiques of published work are submitted through the peer-review system, as psychologists were recently exhorted to do by a senior figure in the field, perhaps we should not be surprised when some people who discover problems in published articles decide to use less formal methods to comment.
(*) Running through this entire post, of course, is the assumption that the reader has set aside for the moment our demonstration of all of the other flaws in the Fredrickson et al. articles, including the massive overfitting and the lack of theoretical coherency. Arguably, those flaws make the entire question of the coding error moot, since even the "corrected" version of the figures very likely fails to correspond to any real effect. But I think it's important to look at this aspect of the story separately from all of the other noise, as an example of how difficult it can be to get even the most obvious errors in the literature corrected.
No comments:
Post a Comment